The Essential Introduction to JupyterLab for Data Science: Your Next-Generation Interactive Computing Hub
The landscape of data science tools is constantly evolving, demanding more integrated, flexible, and powerful environments. While the classic Jupyter Notebook revolutionized interactive computing and data storytelling, its successor, JupyterLab, represents a significant leap forward. It’s not just an upgrade; it’s a fundamental rethinking of the Jupyter experience, designed from the ground up to be the definitive workbench for scientists, engineers, and analysts working with data.
This comprehensive guide serves as your essential introduction to JupyterLab. We’ll delve deep into its architecture, core components, features, and customization options, specifically highlighting why it has become an indispensable tool for modern data science workflows. Whether you’re migrating from the classic Notebook, are new to the Jupyter ecosystem, or simply want to maximize your productivity, this article will equip you with the knowledge to harness the full potential of JupyterLab.
1. What is JupyterLab? The Evolution of Interactive Computing
At its heart, JupyterLab is an interactive development environment (IDE) for computational science, built upon the foundational concepts of the Jupyter Notebook. It provides a web-based user interface that allows you to work with a variety of documents and activities, including:
- Jupyter Notebooks: The familiar
.ipynbfiles for combining code, text, equations, and visualizations. - Code Consoles: Interactive shells for quick code execution and exploration, often linked to notebook kernels.
- Terminals: Full access to your system’s command line directly within the browser.
- Text Editors: Versatile editors with syntax highlighting for various file types (Python scripts, Markdown, JSON, CSV, etc.).
- File Browser: Intuitive navigation and management of your project files and directories.
- Data Viewers: Tools for inspecting common data formats like CSV files.
- Output Views: Dedicated areas for viewing rich outputs generated by notebooks or code.
The key differentiator of JupyterLab is its flexible and modular architecture. Unlike the classic Notebook’s single-document focus, JupyterLab allows you to arrange multiple documents, terminals, consoles, and outputs side-by-side in a customizable layout using tabs and splitters. This transforms it from a simple notebook server into a versatile and integrated workspace.
Key Design Philosophies:
- Integration: Bring diverse tools (notebooks, terminals, editors) into a single, cohesive interface.
- Flexibility: Allow users to arrange their workspace according to their specific needs and workflow.
- Extensibility: Provide a robust extension system enabling the community and third parties to add new features and integrations.
- Modern Web Technologies: Leverage cutting-edge web technologies (like React.js, TypeScript, Lumino) for a responsive and powerful user experience.
2. Why JupyterLab for Data Science? The Compelling Advantages
Data science is inherently iterative and multi-faceted. It involves data exploration, cleaning, modeling, visualization, documentation, and communication. JupyterLab directly addresses the needs of this complex workflow:
- Unified Environment: No more juggling multiple windows for your notebook, terminal, file explorer, and text editor. JupyterLab consolidates these essential tools, reducing context switching and streamlining your process. Need to run a shell command while looking at your data? Open a terminal tab next to your notebook. Need to edit a utility Python script used by your notebook? Open it in another tab.
- Enhanced Productivity through Layout Customization: Arrange your workspace precisely how you like it. View code and its output side-by-side. Compare two notebooks. Monitor a running process in a terminal while editing code. Keep a data file open for reference next to your analysis notebook. This level of customization significantly boosts efficiency.
- Improved Resource Management: The “Running Terminals and Kernels” panel provides a clear overview of active sessions, allowing you to monitor and shut down resources easily, preventing unnecessary memory consumption.
- Seamless Integration with Diverse File Types: JupyterLab isn’t just about notebooks. It handles
.pyscripts, Markdown files, CSVs, JSON, images, and more. You can edit configuration files, write documentation, and inspect data files all within the same interface. - Rich Output Handling: View complex outputs like interactive visualizations (Plotly, Bokeh), geospatial maps (Leaflet), and large data tables more effectively, sometimes in dedicated output views or separate windows.
- Extensibility for Specialized Needs: The powerful extension system allows you to tailor JupyterLab with tools specific to your domain. Popular extensions add features like Git integration, variable inspectors, debuggers, SQL interfaces, language server protocol support (for better code completion and linting), and much more. This transforms JupyterLab from a general tool into a specialized powerhouse.
- Foundation for Collaboration: While real-time collaboration is still evolving, the structured nature of JupyterLab and extensions like Git integration facilitate better teamwork compared to managing standalone classic notebooks.
- Modern and Future-Proof: As the official successor to the classic Notebook, JupyterLab receives active development and represents the future direction of the Jupyter project. Investing time in learning JupyterLab ensures you’re aligned with the ecosystem’s evolution.
JupyterLab vs. Classic Jupyter Notebook:
| Feature | Classic Jupyter Notebook | JupyterLab |
|---|---|---|
| Interface | Single document per browser tab | Multi-document, tabbed, split-view interface |
| Components | Primarily Notebooks, limited Terminal | Notebooks, Consoles, Terminals, Text Editor, File Browser, etc. |
| Layout | Fixed | Flexible, customizable, drag-and-drop |
| Extensibility | Limited (nbextensions) |
Robust, modern extension system (labextensions) |
| File Handling | Basic file navigation, download/upload | Integrated file browser, multi-format editor |
| Architecture | Older, less modular | Modern, modular, extensible (Lumino framework) |
| Development | Maintenance mode | Active development, feature-rich |
| Focus | Document-centric (the Notebook) | Workspace-centric (integrated environment) |
While the classic Notebook remains functional and familiar to many, JupyterLab offers a significantly richer and more productive experience for serious data science work.
3. Getting Started: Installation and Launch
Getting JupyterLab running on your system is straightforward, especially if you’re already using Python environments. The most common methods are via Anaconda/Miniconda or pip.
Method 1: Using Anaconda/Miniconda (Recommended)
Anaconda is a popular Python distribution for data science that includes conda, a powerful package and environment manager. It simplifies the installation of JupyterLab and its dependencies.
- Install Anaconda or Miniconda: If you haven’t already, download and install Anaconda (includes many packages) or Miniconda (minimal installer) from their official websites. Follow the instructions for your operating system.
- Open Your Terminal/Anaconda Prompt:
- Windows: Search for “Anaconda Prompt” and open it.
- macOS/Linux: Open your standard terminal.
- (Optional but Recommended) Create a Dedicated Environment: It’s best practice to create separate environments for different projects to avoid package conflicts.
bash
conda create --name mylabenv python=3.10 # Or your desired Python version
conda activate mylabenv - Install JupyterLab:
bash
conda install -c conda-forge jupyterlab
(Using theconda-forgechannel is often recommended for the latest versions and broader package availability). - Launch JupyterLab: Once the installation is complete, simply run:
bash
jupyter lab
This command will start the JupyterLab server and typically open a new tab in your default web browser pointing to the JupyterLab interface (usuallyhttp://localhost:8888/lab). The terminal where you ran the command will show server logs. Keep this terminal running; closing it will shut down the JupyterLab server.
Method 2: Using pip
If you prefer using pip (Python’s standard package installer), ensure you have Python and pip installed. Using virtual environments is highly recommended here as well.
- Open Your Terminal/Command Prompt.
- (Optional but Recommended) Create and Activate a Virtual Environment:
bash
python -m venv mylabenv # Create environment
# Activate:
# Windows: .\mylabenv\Scripts\activate
# macOS/Linux: source mylabenv/bin/activate - Install JupyterLab using pip:
bash
pip install jupyterlab - Launch JupyterLab:
bash
jupyter lab
Accessing JupyterLab:
Once launched, JupyterLab will open in your browser, displaying the file browser rooted in the directory where you ran the jupyter lab command. You can navigate folders and start creating or opening files.
To stop the server, go back to the terminal where it’s running and press Ctrl+C twice.
4. Exploring the JupyterLab Interface: Anatomy of Your Workspace
The JupyterLab interface is designed to be intuitive yet powerful. Let’s break down its main components:
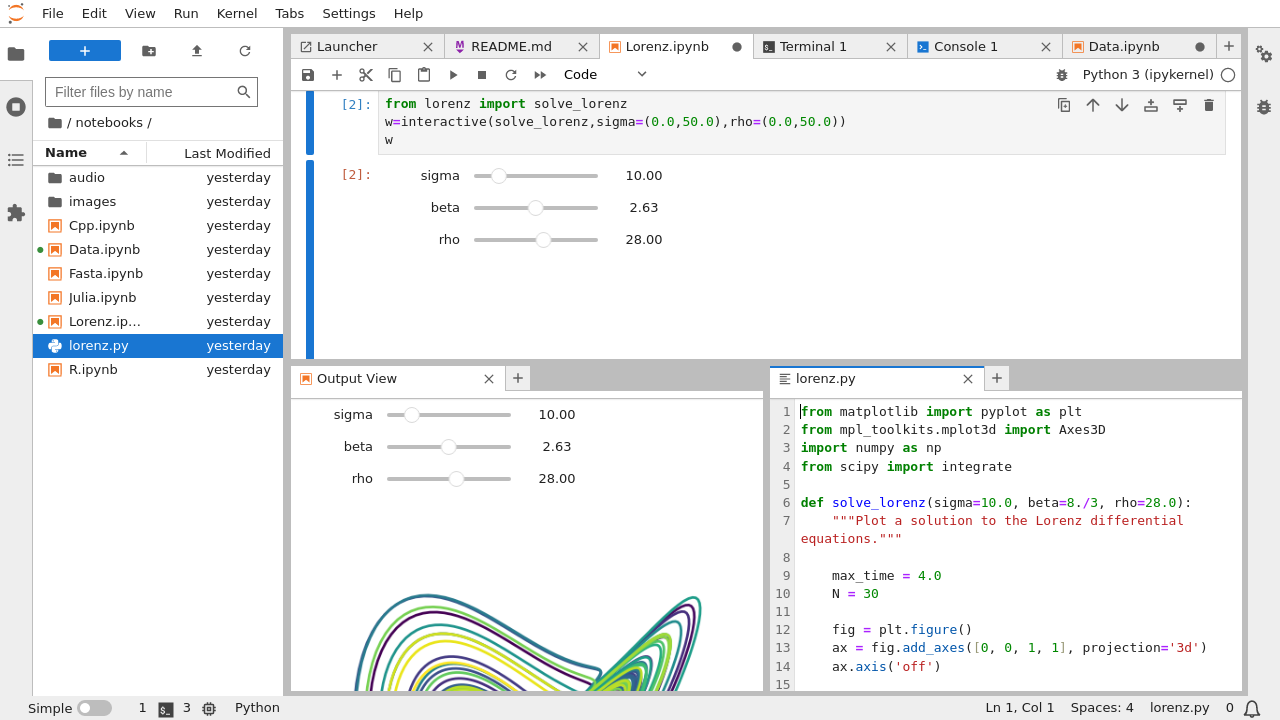 (Note: Actual image display isn’t possible here, but imagine a standard IDE layout)
(Note: Actual image display isn’t possible here, but imagine a standard IDE layout)
-
Menu Bar: Located at the very top, the menu bar contains familiar top-level menus like:
- File: Actions for creating new files (notebooks, consoles, terminals, text files), opening files, saving, closing, exporting, printing, and quitting.
- Edit: Standard editing commands (undo, redo, cut, copy, paste), plus cell-specific operations (split cell, merge cells), find/replace, and clearing outputs.
- View: Controls for toggling the visibility of sidebars, status bar, changing themes (Light/Dark), controlling notebook views (single/multiple document mode), activating presentation mode, and managing text editor settings (line numbers, wrap words).
- Run: Commands for executing code cells in notebooks or consoles (Run Selected Cells, Run All Cells, Restart Kernel and Run All).
- Kernel: Actions for managing the execution kernels associated with notebooks and consoles (Interrupt Kernel, Restart Kernel, Shut Down Kernel, Change Kernel).
- Tabs: Navigation and management of open tabs within the main work area.
- Settings: Access to theme selection, text editor settings, keyboard shortcuts customization, and the Advanced Settings Editor (for fine-grained JSON-based configuration).
- Help: Links to JupyterLab documentation, reference materials, kernel help, and information about installed extensions.
-
Left Sidebar: A collapsible panel on the left containing several tabs for key functionalities:
- File Browser (
Folder Icon): Navigate the file system, create new folders, upload/download files, rename, delete, and open files in the main work area. This is your primary way to manage project assets. - Running Terminals and Kernels (
Terminal/Power Icon): Lists all active kernel sessions (for notebooks and consoles) and running terminal sessions. You can see which files are using which kernels and shut them down individually or all at once. Essential for resource management. - Commands (
Palette Icon): A searchable palette of all available commands in JupyterLab, including those from extensions. A quick way to find and execute actions without memorizing menus or shortcuts. - Table of Contents (
List Icon): Automatically generates an interactive table of contents based on the Markdown headings in the currently active notebook or Markdown file. Great for navigating long documents. - Extension Manager (
Puzzle Piece Icon): Discover, install, uninstall, enable, and disable JupyterLab extensions. This is the gateway to customizing and extending JupyterLab’s capabilities. (Note: May require Node.js installation and enabling via settings).
- File Browser (
-
Main Work Area: This is the central, largest part of the interface where your documents and activities live.
- Tabs: Open documents (notebooks, text files, terminals, etc.) appear as tabs across the top of this area.
- Panels/Splitters: You can drag tabs to the sides or bottom of other tabs to create split views. This allows you to arrange multiple documents side-by-side or top-and-bottom. You can resize these panels by dragging the splitters between them. This flexible layout is a core feature of JupyterLab.
-
Status Bar: Located at the bottom, providing contextual information about the current state:
- Kernel status (Idle, Busy, Connecting, etc.) for the active notebook/console.
- Current Python environment (if configured).
- Cursor position and indentation settings in text editors/code cells.
- Other status messages from extensions.
Understanding these core areas allows you to navigate and utilize the JupyterLab environment effectively. Spend some time clicking around, opening different file types, and experimenting with dragging tabs to create different layouts.
5. Core Components and Concepts In-Depth
Let’s dive deeper into the essential building blocks you’ll interact with constantly in JupyterLab.
a) Jupyter Notebooks (.ipynb)
The heart of interactive computing in Jupyter.
* Structure: Composed of individual cells. Cells can contain Code, Markdown (for formatted text, equations, images), or Raw content.
* Kernels: Each notebook is connected to a kernel, which is the computational engine that executes the code within the notebook’s code cells. The default kernel is usually IPython (for Python), but many other kernels exist for languages like R, Julia, Scala, etc. You select the kernel when creating a new notebook or can change it later via the Kernel menu.
* Execution Flow: Code cells are executed sequentially (though you can run them out of order). The output of a code cell (text, plots, tables) appears directly below it. The state (variables, functions defined) persists within the kernel session until it’s restarted.
* Rich Output: Notebooks excel at displaying rich outputs, including static plots (Matplotlib, Seaborn), interactive visualizations (Plotly, Bokeh, Altair), HTML tables (like pandas DataFrames), LaTeX equations, images, and videos.
b) Code Consoles
Think of these as interactive REPLs (Read-Eval-Print Loops) within JupyterLab.
* Purpose: Ideal for quick, exploratory coding, testing snippets, or interacting with the kernel of an existing notebook without cluttering the notebook itself.
* Linking: You can create a console linked to a notebook’s kernel. This means the console shares the same execution state (variables, imported modules) as the notebook. Changes made in one affect the other.
* Use Cases: Quickly check a variable’s value, test a function, try out a library call before adding it to your main notebook workflow.
c) Terminals
Provides direct access to your system’s shell (Bash, Zsh, PowerShell, etc.) within a JupyterLab tab.
* Functionality: Execute any command-line tool, manage files, install packages (using pip or conda if within the correct environment), run scripts, use version control (Git), monitor system resources.
* Integration: Invaluable for tasks that complement your data science workflow without leaving the JupyterLab environment. For instance, downloading data using curl or wget, managing Conda environments, or running external processing scripts.
d) Text Editor
A capable editor for various plain text file formats.
* Features: Syntax highlighting for numerous languages (Python, R, SQL, Markdown, JSON, YAML, HTML, CSS, JavaScript, etc.), configurable indentation, themes (matching JupyterLab’s theme), find/replace, line numbers, word wrap.
* Use Cases: Editing Python utility scripts (.py) that your notebooks import, writing Markdown documentation (.md), viewing and editing configuration files (.json, .yaml), inspecting data files (.csv, .tsv), or even light web development (.html, .css).
e) File Browser
Your window into the project’s directory structure.
* Navigation: Browse folders and files on the server where JupyterLab is running.
* File Operations: Create new folders, text files, notebooks; upload files from your local machine; download files/folders from the server; rename, delete, cut, copy, paste files and folders.
* Context Menu: Right-clicking on files/folders provides relevant actions (Open With…, Rename, Delete, Duplicate, Copy Path, etc.).
f) Kernels Explained
Kernels are the unsung heroes making interactive computing possible.
* Role: A kernel is a separate process that runs your code in a specific language. JupyterLab communicates with the kernel, sending code from cells for execution and receiving back the results (output, errors) to display.
* Language Support: While IPython is the default Python kernel, Jupyter supports kernels for dozens of languages (R, Julia, Scala, Bash, SQL, etc.), making JupyterLab a polyglot environment. You need to install the respective kernel separately (e.g., IRkernel for R).
* Isolation: Each notebook or console typically runs in its own kernel process. This isolates their execution environments, preventing variables or imports in one notebook from interfering with another (unless explicitly designed to share via consoles).
* Management: The “Running Kernels” tab and the Kernel menu allow you to see active kernels, shut them down to free resources, restart them if they become unresponsive or if you want a clean state, or change the kernel associated with a notebook. Regularly restarting kernels is good practice to ensure reproducibility.
g) Cells: The Building Blocks of Notebooks
- Code Cells: Contain code to be executed by the notebook’s kernel. Identified by
In [ ]:to the left. PressShift+Enter(run and select next cell) orCtrl+Enter(run and stay on current cell) to execute. - Markdown Cells: Contain text formatted using Markdown syntax. Used for documentation, explanations, section headings, embedding images, and writing LaTeX equations (rendered beautifully using MathJax). Double-click to edit,
Shift+EnterorCtrl+Enterto render. - Raw NBConvert Cells: Content in these cells is passed through unmodified when converting the notebook to other formats using
nbconvert. Less commonly used for direct analysis.
Understanding these components and how they interact is crucial for leveraging the power and flexibility of JupyterLab.
6. Core JupyterLab Features for Data Science Workflows
Now let’s focus on how JupyterLab’s features specifically benefit common data science tasks.
a) Interactive Computing and Exploration
This is the bedrock of Jupyter. The notebook format encourages an iterative cycle:
1. Write Code: Input Python code (e.g., using pandas) to load or manipulate data in a cell.
2. Execute: Run the cell (Shift+Enter).
3. Inspect Output: Immediately see the results (DataFrame preview, variable values, plot).
4. Refine: Modify the code or write new code in the next cell based on the output.
JupyterLab enhances this with:
* Code Consoles: Quickly test variations of code or inspect intermediate states without adding temporary cells to your main notebook.
* Side-by-Side Views: View your data (e.g., in a CSV viewer extension or as DataFrame output) next to the code manipulating it.
b) Code Execution and Debugging
- Kernel Management: Easily restart kernels for a clean slate or interrupt long-running computations. Change kernels if you need to switch between, say, a standard Python environment and one with specialized deep learning libraries.
- Debugging (via Extensions): While basic
print()debugging works, extensions likejupyterlab-debuggerprovide a visual debugger experience (setting breakpoints, stepping through code, inspecting variables) directly within the notebook interface, significantly improving the debugging process.
c) Documentation and Storytelling with Markdown
Data science isn’t just about code; it’s about explaining the process and results.
* Rich Text: Use Markdown cells to structure your analysis with headings, lists, bold/italic text, links, and blockquotes.
* Mathematical Equations: Embed complex mathematical formulas and equations using LaTeX syntax ($inline$, $$display$$).
* Embedding Images: Include diagrams, logos, or static result images directly in your narrative.
* Table of Contents: The built-in ToC generator makes navigating long, well-structured notebooks effortless.
d) Data Visualization
Visualizations are critical for understanding data and communicating findings.
* Inline Plotting: Standard libraries like Matplotlib and Seaborn render plots directly within the notebook output cells.
“`python
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
# Example Data
data = pd.DataFrame({'x': np.arange(10), 'y': np.random.rand(10)*10})
# Matplotlib Plot
plt.figure()
plt.plot(data['x'], data['y'], marker='o')
plt.title("Simple Matplotlib Plot")
plt.xlabel("X-axis")
plt.ylabel("Y-axis")
plt.show() # Renders inline
# Seaborn Plot
plt.figure()
sns.scatterplot(data=data, x='x', y='y')
plt.title("Simple Seaborn Plot")
plt.show() # Renders inline
```
- Interactive Visualizations: Libraries like Plotly, Bokeh, and Altair generate interactive plots (zooming, panning, hovering) that remain live within the notebook or can be opened in separate views. JupyterLab’s architecture handles these rich outputs smoothly.
- Layout Flexibility: View complex plots alongside the code that generated them, or even compare multiple plots side-by-side in different panels.
e) Data Exploration and Inspection
-
Pandas Integration: The primary tool for data manipulation in Python, pandas DataFrames, render as nicely formatted HTML tables in notebook outputs.
“`python
import pandas as pd
import numpy as npCreate a sample DataFrame
df = pd.DataFrame({
‘A’: np.random.randn(5),
‘B’: pd.date_range(‘20230101’, periods=5),
‘C’: pd.Categorical([‘test’, ‘train’, ‘test’, ‘train’, ‘test’]),
‘D’: np.random.randint(0, 100, size=5)
})print(“DataFrame Info:”)
df.info() # Prints summary infoprint(“\nDataFrame Head:”)
display(df.head()) # Renders HTML table in output
``lckr-jupyterlab-variableinspector` provide a panel showing currently defined variables in the kernel, their types, sizes, and values, offering a great way to track the state of your workspace.
* **CSV/Data Viewers:** Open CSV, TSV (and potentially other formats via extensions) in a spreadsheet-like viewer within a JupyterLab tab for quick inspection without loading them into memory with pandas.
* **Variable Inspectors (via Extensions):** Extensions like
7. Advanced Features and Customization
JupyterLab truly shines when you start leveraging its more advanced capabilities and tailoring it to your preferences.
a) Workspace and Layout Management
- Drag-and-Drop: Drag tabs to the edges of panels or other tabs to create splits (horizontal or vertical). Rearrange tabs within a panel.
- Multiple Views of the Same Document: Right-click a notebook tab and select “New View for Notebook”. You can have multiple synchronized views of the same notebook, perhaps looking at the code in one view and the rendered Markdown or plots in another.
- Simple Interface Mode: For a less cluttered view focusing on a single document (similar to the classic Notebook), use
View -> Simple Interface. - Saving Layouts (Implicit): JupyterLab attempts to restore your previous session’s layout (open tabs and their arrangement) when you restart it, though this behavior can sometimes be inconsistent depending on the shutdown process. Explicit layout saving might come in future versions or via extensions.
b) Themes
Easily switch between predefined light and dark themes.
* Access: Settings -> Theme -> JupyterLab Light / JupyterLab Dark.
* Custom Themes: Many community-created themes can be installed as extensions, allowing you to personalize the look and feel further. Search the Extension Manager for “theme”.
c) Keyboard Shortcuts
Mastering keyboard shortcuts dramatically speeds up your workflow. JupyterLab has two main modes for notebooks:
* Command Mode (Blue Left Border): Press Esc to enter. Shortcuts affect the notebook structure (e.g., A to insert cell above, B below, DD delete cell, M change to Markdown, Y change to Code).
* Edit Mode (Green Left Border): Press Enter or click inside a cell. Standard text editing shortcuts apply, plus Ctrl+Enter (run cell) and Shift+Enter (run cell, select below).
Key Default Shortcuts (Command Mode):
Shift+Enter: Run cell, select belowCtrl+Enter: Run selected cellsAlt+Enter: Run cell, insert belowA: Insert cell aboveB: Insert cell belowX: Cut selected cellsC: Copy selected cellsV: Paste cells belowShift+V: Paste cells aboveD, D(press twice): Delete selected cellsZ: Undo cell deletionM: Change cell type to MarkdownY: Change cell type to CodeL: Toggle line numbers in cellShift+L: Toggle line numbers in all cellsCtrl+Shift+C: Open Command PaletteCtrl+Shift+F: Open File Browser in left sidebarCtrl+B: Toggle Left Sidebar visibility
Customization: Access Settings -> Advanced Settings Editor -> Keyboard Shortcuts to view and modify shortcuts using JSON overrides.
d) The Power of Extensions
Extensions are third-party additions that significantly enhance JupyterLab’s functionality. They are the key to tailoring the environment to specific needs.
- Finding and Installing: Use the Extension Manager tab (puzzle piece icon) in the left sidebar. You can search for available extensions, install, uninstall, enable, or disable them. Installation often requires Node.js to be installed on the server machine and might necessitate rebuilding JupyterLab (
jupyter lab build), though newer versions handle this more gracefully. Be cautious: Installing many or conflicting extensions can sometimes lead to instability. - Essential Extensions for Data Science:
jupyterlab-git: Adds a Git panel to the left sidebar for managing repositories (staging, committing, pushing, pulling, branching, viewing history) directly within JupyterLab. Indispensable for version control.jupyterlab-lsp(Language Server Protocol): Integrates language servers (e.g.,python-lsp-server) to provide advanced coding features like intelligent code completion, linting (error/style checking), jump-to-definition, renaming, and diagnostics within notebooks and the text editor. Requires installing the language server itself (pip install 'python-lsp-server[all]'). Transforms the coding experience.jupyterlab-debugger: Provides a visual debugger interface for IPython kernels (requires thexeus-pythonkernel oripykernel>=6.0). Set breakpoints, step through code, inspect variables.jupyterlab-variableInspector: Displays a list of active variables, their types, sizes, and values in a sidebar tab. Helps track kernel state.@jupyterlab/toc: (Often built-in now) The Table of Contents generator.jupyterlab-plotly/@bokeh/jupyter_bokeh: Ensure interactive plots from Plotly or Bokeh render correctly and efficiently within JupyterLab.jupyter-server-proxy: Allows running and accessing other web applications (like TensorBoard, MLflow UI, Streamlit apps) through the JupyterLab interface.jupyterlab_execute_time: Displays the execution time and timestamp for each code cell. Useful for performance monitoring.jupyterlab-drawio: Embed and edit Draw.io diagrams within JupyterLab. Great for workflow visualization.- SQL Extensions (e.g.,
jupyterlab-sql): Add interfaces for connecting to databases, browsing schemas, and running SQL queries directly within Lab.
Managing and selecting the right extensions can turn your JupyterLab instance into a highly specialized and efficient data science workbench.
8. A Typical Data Science Workflow Example in JupyterLab
Let’s walk through a simplified data science task to illustrate how JupyterLab’s components work together. Imagine we have a G CSV file (data.csv) containing some sales data.
-
Setup:
- Launch JupyterLab from your project directory.
- Use the File Browser (left sidebar) to confirm
data.csvis present or upload it. - Create a new Notebook (
File -> New -> Notebook, select Python 3 kernel). Rename itSales_Analysis.ipynb.
-
Loading Data (Notebook):
-
In the first Code Cell, import pandas and load the data:
“`python
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as snsprint(“Loading data…”)
try:
df = pd.read_csv(‘data.csv’)
print(“Data loaded successfully.”)
except FileNotFoundError:
print(“Error: data.csv not found in the current directory.”)
# Optionally, create dummy data for demonstration
df = pd.DataFrame({
‘Date’: pd.to_datetime([‘2023-01-15’, ‘2023-01-16’, ‘2023-02-10’, ‘2023-02-11’, ‘2023-03-05’]),
‘Product’: [‘A’, ‘B’, ‘A’, ‘C’, ‘B’],
‘Sales’: [100, 150, 120, 80, 200],
‘Region’: [‘North’, ‘South’, ‘North’, ‘West’, ‘South’]
})
print(“Created dummy data.”)Display first few rows
display(df.head())
``Shift+Enter
* Run the cell (). The output will show the print statements and the HTML table fordf.head()`.
-
-
Initial Exploration (Notebook & Console):
-
In the next Code Cell:
“`python
print(“Data Info:”)
df.info()print(“\nDescriptive Statistics:”)
display(df.describe())print(“\nValue Counts for Product:”)
display(df[‘Product’].value_counts())
``File -> New -> Console
* Run this cell.
* **Scenario:** You want to quickly check the data type of the 'Date' column *without* adding another cell.
* Go to. Select the kernel associated withSales_Analysis.ipynb.df[‘Date’].dtype
* In the **Console**, typeand pressShift+Enter. The output (dtype(‘<M8[ns]’)` or similar for datetime) appears instantly. This confirms the data type without modifying the notebook structure.
-
-
Data Cleaning / Feature Engineering (Notebook & Text Editor):
- Scenario: Let’s say you need a reusable function to extract the month from the date. You decide to put it in a separate utility script.
- Go to
File -> New -> Text File. Rename itutils.py. -
In the Text Editor tab for
utils.py, write the function:
“`python
# utils.py
import pandas as pddef get_month(date_series):
“””Extracts the month from a pandas Series of datetime objects.”””
if pd.api.types.is_datetime64_any_dtype(date_series):
return date_series.dt.month
else:
# Attempt conversion if not already datetime
try:
return pd.to_datetime(date_series).dt.month
except Exception as e:
print(f”Error converting to datetime: {e}”)
return None
* Save `utils.py` (`Ctrl+S`).python
* Go back to your `Sales_Analysis.ipynb` tab.
* In a new **Code Cell**, import and use the function:
import utils # Import our utility scriptdf[‘Month’] = utils.get_month(df[‘Date’])
print(“Added ‘Month’ column:”)
display(df.head())
``Sales_Analysis.ipynb
* Run the cell. You can haveandutils.py` open side-by-side using drag-and-drop for easy reference.
-
Visualization (Notebook):
-
In a new Code Cell, create some plots:
“`python
plt.figure(figsize=(10, 5))plt.subplot(1, 2, 1) # 1 row, 2 cols, plot 1
sns.histplot(data=df, x=’Sales’, kde=True)
plt.title(‘Sales Distribution’)plt.subplot(1, 2, 2) # 1 row, 2 cols, plot 2
monthly_sales = df.groupby(‘Month’)[‘Sales’].sum()
monthly_sales.plot(kind=’bar’)
plt.title(‘Total Sales by Month’)
plt.xlabel(‘Month’)
plt.ylabel(‘Total Sales’)
plt.xticks(rotation=0)plt.tight_layout() # Adjust spacing
plt.show()
“`
* Run the cell. The plots will appear inline.
-
-
Documentation (Notebook):
- Insert Markdown Cells between code cells to explain the steps:
- Above the loading cell:
## 1. Load Sales Data - Above exploration:
## 2. Initial Data Exploration - Above cleaning:
## 3. Data Cleaning and Feature Engineering \n We add a 'Month' column using a function fromutils.pyfor better analysis. - Above visualization:
## 4. Visualize Sales Patterns
- Above the loading cell:
- Render the Markdown cells (
Shift+Enter). Use the Table of Contents tab in the left sidebar to quickly navigate your analysis.
- Insert Markdown Cells between code cells to explain the steps:
-
Version Control (Git Extension):
- Assuming you initialized a Git repository in your project directory (
git initin the terminal or via the extension). - Open the Git tab in the left sidebar (puzzle piece icon, requires
jupyterlab-gitextension). - See your changed files (
Sales_Analysis.ipynb,utils.py). - Stage the changes (click
+next to files). - Enter a commit message (e.g., “Initial sales analysis and plotting”).
- Click the Commit button.
- Assuming you initialized a Git repository in your project directory (
This example demonstrates how notebooks, consoles, text editors, the file browser, visualization, Markdown, and extensions (like Git) come together in JupyterLab to facilitate a fluid data science workflow.
9. Best Practices and Tips for Effective JupyterLab Usage
- Environment Management: Always use dedicated virtual environments (conda or venv) for your projects to isolate dependencies. Activate the correct environment before launching
jupyter lab. - Kernel Management: Restart kernels frequently (
Kernel -> Restart Kernel...or the refresh icon) to ensure your code runs correctly from top to bottom and doesn’t depend on hidden state from previous executions. Shut down unused kernels using the “Running” tab to save resources. - Modular Code: For complex or reusable logic, move code from notebooks into separate Python scripts (
.pyfiles) and import them, as shown in the workflow example. This improves organization, testability, and reusability. - Notebook Structure: Use Markdown headings (
#,##,###) to structure your notebooks logically. Keep notebooks focused on a specific task or analysis. Split very long analyses into multiple notebooks. - Cell Granularity: Break down code into logical cells. Avoid overly long cells. Each cell should ideally perform a distinct step.
- Version Control: Use Git religiously. Commit frequently with meaningful messages. The
jupyterlab-gitextension makes this seamless. Configure.gitignoreto exclude large data files and environment directories (likevenv/or__pycache__/). Note: Diffing notebooks in Git can be tricky due to output and metadata; tools likenbdimecan help. - Documentation: Write clear Markdown explanations for your code, assumptions, and findings. Your future self (and collaborators) will thank you.
- Naming Conventions: Use clear and consistent names for notebooks, files, variables, and functions.
- Resource Monitoring: Keep an eye on the “Running” tab. If your machine becomes sluggish, check for and shut down unnecessary kernels or terminals.
- Explore Extensions: Periodically browse the Extension Manager for tools that could improve your specific workflow (debugging, linting, data viewing, etc.).
- Keyboard Shortcuts: Invest time in learning the key shortcuts for navigation, cell execution, and manipulation. It pays off significantly in speed.
- Clean Outputs: Before saving or committing a final version of a notebook, consider using
Kernel -> Restart Kernel and Run All Cells...for a clean execution trail, orEdit -> Clear All Outputsif you only want to share the code and Markdown.
10. Troubleshooting Common Issues
- Kernel Won’t Start / Kernel Dying:
- Ensure the kernel’s dependencies (like
ipykernel) are installed in the correct Python environment from which you launched JupyterLab. - Check for resource exhaustion (CPU/Memory). Shut down other kernels/processes.
- Look for errors in the terminal where
jupyter labis running. - Try creating a fresh environment and reinstalling core packages.
- A specific library call might be crashing the kernel; try commenting out recent code additions.
- Ensure the kernel’s dependencies (like
ModuleNotFoundError:- The required package is likely not installed in the active kernel’s environment. Open a Terminal within JupyterLab (
File -> New -> Terminal) and install the package (pip install <package_name>orconda install <package_name>if using conda environment). You might need to restart the kernel afterwards. - Ensure you launched
jupyter labfrom the terminal where the correct environment is activated.
- The required package is likely not installed in the active kernel’s environment. Open a Terminal within JupyterLab (
- JupyterLab Slow or Unresponsive:
- Too many active kernels or browser tabs. Close unused ones.
- Very large notebooks or notebooks with massive outputs (huge DataFrames, complex interactive plots). Clear outputs (
Edit -> Clear All Outputs) or break down the notebook. - Browser issues: Try clearing browser cache, disabling browser extensions, or using a different browser.
- Extension conflicts: Try disabling recently installed or potentially problematic JupyterLab extensions via the Extension Manager.
- Server resource limits: The machine running JupyterLab might be underpowered.
- Extension Installation Problems:
- Ensure you have Node.js installed (required for building many extensions).
- Check for compatibility issues between extensions or with your JupyterLab version.
- Look for error messages during the
jupyter lab buildprocess (if required). Consult the specific extension’s documentation.
- Websocket Errors / Connection Issues:
- Often related to network configuration, proxies, or firewalls.
- Check the terminal logs for detailed error messages.
- Try accessing JupyterLab using
127.0.0.1instead oflocalhostin the URL.
11. The Future of JupyterLab and Conclusion
JupyterLab is not a static product; it’s an actively developed open-source project with a vibrant community. Future developments likely include:
- Enhanced Real-time Collaboration: Improving the experience for multiple users working on the same documents simultaneously.
- More Core Extensions: Integrating more popular functionalities directly into the base distribution.
- Improved Accessibility: Continuing efforts to make the interface usable for everyone.
- Performance Optimizations: Further refining the performance, especially with large notebooks and datasets.
- Tighter IDE Integration: Blurring the lines further between a notebook environment and a full-fledged IDE, potentially with more sophisticated refactoring tools and debugging capabilities.
- JupyterLab Desktop App: A standalone desktop application wrapping the web interface, simplifying installation and integration.
Conclusion:
JupyterLab represents a paradigm shift in interactive computing for data science. By providing a flexible, integrated, and extensible environment, it moves beyond the single-document focus of the classic Notebook to become a comprehensive workbench. Its ability to seamlessly combine notebooks, code editors, terminals, consoles, and rich outputs within a customizable layout directly addresses the multifaceted nature of the data science workflow.
While the classic Notebook laid the foundation, JupyterLab builds upon it with modern web technologies, a robust extension ecosystem, and a user experience designed for productivity. Mastering JupyterLab empowers data scientists to explore data, develop models, visualize results, and document their work more effectively and efficiently than ever before. If you’re serious about data science in the Python ecosystem (and beyond), investing time to learn and adopt JupyterLab is not just beneficial – it’s essential for staying at the forefront of modern computational science. Start exploring, customize your workspace, leverage extensions, and unlock a new level of productivity in your data journey.